Successfully edited the genes of intestinal bacteria in living mice
A research team led by Xavier Duportet, a synthetic biologist and co-founder of biotech company Eligo Bioscience, has successfully designed a gene editing tool that can modify bacterial populations in the microbiome. intestinal flora of live mice.
Initial experimental results showed that this tool helped modify targeted genes in more than 90% of Escherichia coli colonies inside the intestinal tract of living mice. Escherichia coli is a type of bacteria that commonly lives in the intestines of humans and animals. Most types of E. coli bacteria are known to cause temporary and transient diarrhea, or some severe intestinal infections lead to more severe illness with diarrhea, abdominal pain and fever.
Some previous research has used the CRISPR—Cas gene editing system to kill harmful bacteria in the intestines of mice. But Duportet and colleagues just wanted to edit the genes of bacteria in the gut microbiome without killing them.
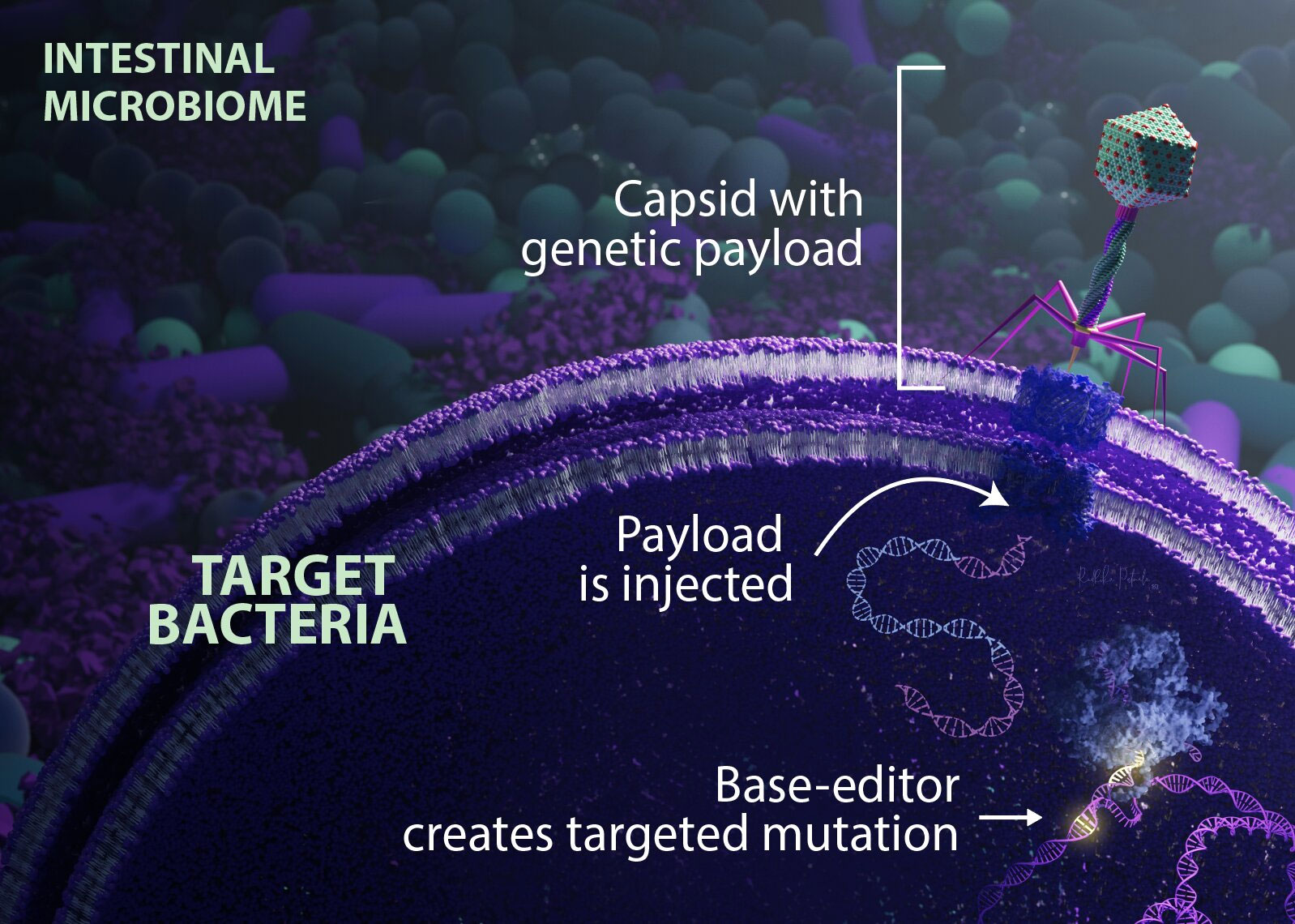
To do this, scientists used a method of swapping one nucleotide base for another - such as converting A to G - without breaking the DNA double strand. To date, most existing methods are unable to sufficiently modify the target bacterial population to be effective. This is because the injected vectors only target target receptors commonly found in bacteria grown in the laboratory.
To overcome this obstacle, Duportet and his colleagues engineered a delivery vehicle that uses components of a bacteriophage - a type of virus that infects bacteria - to contain some of the receptors E. coli expresses in the intestinal environment. This vector carries a 'basic gene editing tool' that targets specific E. coli genes. The team also improved the system to prevent the provided genetic material from replicating and spreading while it is inside the bacteria.
The team inserted the basic gene editing tool into mice and used it to change A to G in the E. coli gene that produces β-lactamase - an enzyme that promotes bacterial resistance to certain antibiotics. About eight hours after the animals were treated, about 93% of the target bacteria had been genetically edited.
The researchers then tweaked the editing tool so that it could modify the E. coli gene to produce a protein believed to play a role in some neurodegenerative and autoimmune diseases. The percentage of edited bacteria hovered around 70% within three weeks after the mice were treated. In the lab, scientists can also use the tool to edit strains of E. coli and Klebsiella pneumoniae that can cause pneumonia infections. This suggests that the editing system can be tailored to target different bacterial strains and species.
The achievement represents a 'major leap' in the development of tools that can transform bacteria directly within the gut, opening up the ability to fight disease, while also blocking harmful DNA from spreading. orchid.
The next step for Duportet and colleagues is to develop mouse models of microbiome-driven diseases to measure whether specific gene editing has beneficial effects on their health.
You should read it
- ★ Scientists generate electricity from the air by nanowires generated from bacteria
- ★ The world of microbes shimmering and fantasy on your teeth
- ★ Finding new gene therapy can help treat hearing loss
- ★ How to teach children how to use the mouse?
- ★ Get fever with new technology to identify E-coli bacteria